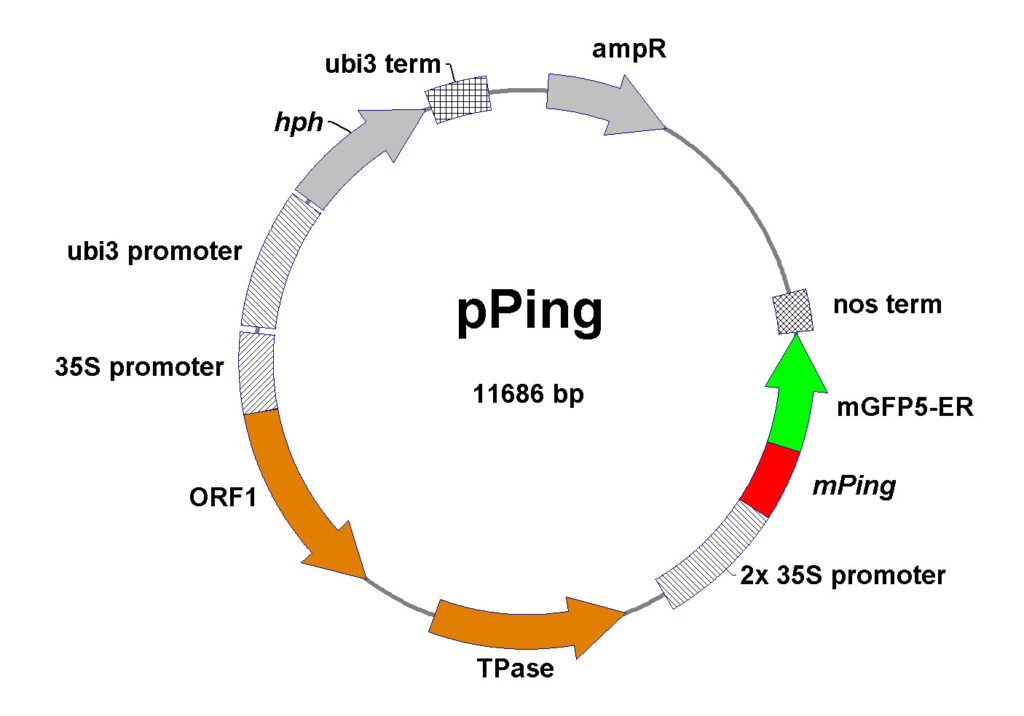
Map of original pPing vector used for Soybean transformation
GFP fluorescence indicating excision of mPing
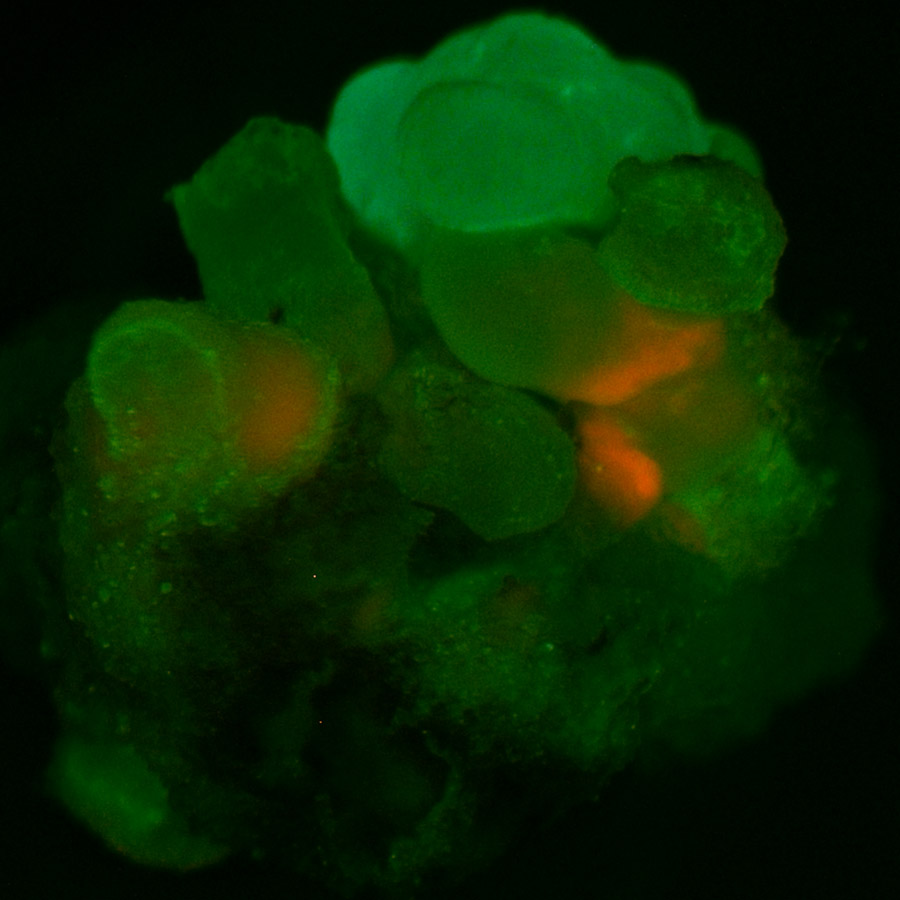
GFP florescence indicating excision of mPing in a sector of soybean embryogenic tissue transformed with the pPing construct (MSD20 + IFT plates).
mPing is a miniature inverted terminal repeat element (MITE) from rice. mPing transposes actively in both anther culture and in some Japanese rice cultivars. Like other members of the Tourist-like class of MITEs, it inserts preferentially in and around genes and can be amplified to high copy numbers (>1000). Inserted elements are flanked by a TAA trinucleotide target site duplication and generally excise precisely. mPing was adapted for use in Arabidopsis, where it was shown to require two open reading frames (ORFs) (ORF1 and TPase) from a full length, autonomous element (either the Ping or Pong) for transposition. mPing was shown to transpose to unlinked sites on all Arabidopsis chromosomes.
The use of mPing as a mutagen in soybean is primarily being performed by the Parrott lab. Results with our original vector show that mPing is capable of transposing and inducing mutant phenotypes in soybean (Hancock et al. 2011)1. Since mPing has proven to be an effective insertional mutagen in soybean we have expanded our work to include both silencing and activation tag verions of mPing.
The goals of the mPing project are:
- Re-design insertional, silencing, and activation tag versions of mPing with a high expressing soybean Ubiquitin promoter
- Increase the transposition rate by placing lines with ORF1 and transposase into tissue culture. 350 lines will be put into tissue culture each year.
- Use and improve a mPing Illumina protocol so that mPing transposition events can be efficiently located within the soybean genome.
Multiple labs have established segregating populations derived from a cross between a mutant mPing line and a non-transgenic control. The resulting populations are being evaluated for segregation of the mutant phenotype. Candidate genes identified from Illumina sequencing of control and mutant plants are being assayed for co-segregation with the mutant phenotype to determine if they are causitive. CRISPR-Cas9 will be employed to validate the genotype/phenotype relationship for agriculturally relevant mutants.
Since mPing transposes to unlinked sites in soybean with nearly 70% of insertions occurring near genes (Hancock et al. 2011)1, the ability of mPing to carry an activation tag element is being evaluated. Activation tagging is typically used to induce misexpression mutations, useful for the identification of repetitive and lethal genes. The original activation tag included two copies (2X) of cassava mosaic virus (CsMV) enhancer sequence embedded within the mPing element. Transposition was confirmed in Arabidopsis before subsequent transformation into soybean. New versions of mPing based activation tags including a non-viral version are currently being tested in hairy roots. The most promising candidates, as assayed by transposition in hairy roots, will be biolistically transformed in cultivars for whole plant evaluation.
An Illumina library protocol was developed to sequence mPing adjacent sequences within the soybean genome. However, many putative insertions identified by Illumina sequences have been deemed false positives by PCR (Kanizay et al., 2015). Modifications to the protocol are being made to enrich for ‘real’ insertions which will allow an increase in the number of individuals successfully pooled into a sequencing run. Additionally, this protocol will be leveraged to map Tnt1 and Ac/Ds transposition events.



mPing mutant phenotypes: rugose, brown seeds and early maturing respectively.
